G2PMineR
A genome to phenome literature review approach
John M.A. Wojahn, Stephanie J. Galla, Anthony E. Melton, Sven Buerki
2021-08-12
1 Why use G2PMineR?
There is a gap in the conceptual framework linking genes to phenotypes (G2P) for non-model or-ganisms, as most non-model organisms do not yet have genomic resources readily available. To address this, researchers often perform literature reviews to understand G2P linkages by curating a list of likely gene candidates, hinging upon other studies already conducted in closely related systems. Sifting through hundreds to thousands of articles is a cumbersome task that slows down the scientific process and may introduce bias into a study. To fill this gap, we created G2PMineR, a free and open-source literature mining tool developed specifically for G2P research. This package uses automation to make the G2P review process efficient and unbiased, while also generating hypothesized associations between genes and phenotypes within a taxonomical framework. We applied the package to a literature review for drought-tolerance in plants. The analysis provides biologically meaningful the results within the known framework of drought tolerance in plants. Overall, the package is useful for conducting literature reviews for ge-nome-to-phenome projects and also has broad appeal to scientists investigating a wide range of study systems as it can conduct analyses under the auspices of three different kingdoms (Plantae, Animalia, and Fungi).
2 Link to Publication
3 Overview
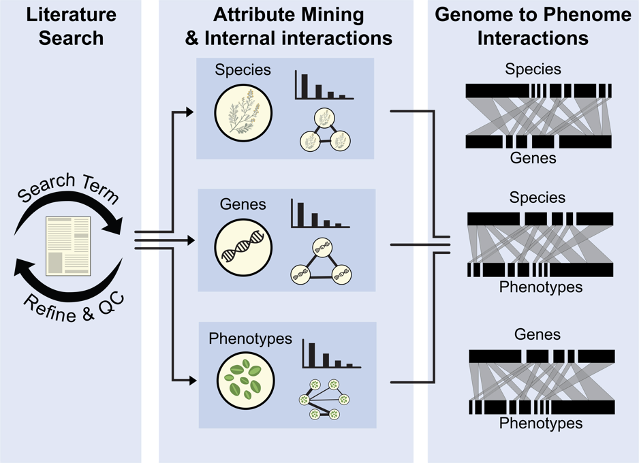
3.1 Steps of a G2PMineR analysis
The G2PMineR package is composed of 3 steps and eight modules. Step 1 and step 2 module 5 are optional:
Step 1: Literature search
- Module 1: Conduct literature search and assess its efficiency.
Step 2: Mining (incl. quality controls) G2P data in abstracts using reference libraries.
- Module 2: Mining taxonomy (Ta).
- Module 3: Mining genes (G).
- Module 4: Mining phenotypes (P).
- Module 5: Summarize and consensus G, Ta and P data.
- Module 6: Internal network analyses for G, Ta and P data.
Step 3: Genome to phenome interactions (rooted into taxonomical framework)
- Module 7: Infer bipartite graphs to link G, Ta and P data.
3.2 G2PMineR Flowchart
[ ]
]
4 Installing G2PMineR
4.1 Dependencies
devtools
You can find G2PMineR on GitHub at wojahn/G2PMineR.
# First install devtools
install.packages("devtools")
# Then install G2PMineR from GitHub
devtools::install_github("BuerkiLabTeam/G2PMineR")5 Input/Output Table
7 Appendix 1
Citations of all R packages used to generate this report.
[1] J. Allaire, Y. Xie, J. McPherson, et al. rmarkdown: Dynamic Documents for R. R package version 2.9. 2021. <URL: https://CRAN.R-project.org/package=rmarkdown>.
[2] C. Boettiger. knitcitations: Citations for Knitr Markdown Files. R package version 1.0.12. 2021. <URL: https://github.com/cboettig/knitcitations>.
[3] M. C. Koohafkan. kfigr: Integrated Code Chunk Anchoring and Referencing for R Markdown Documents. R package version 1.2. 2015. <URL: https://github.com/mkoohafkan/kfigr>.
[4] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2020. <URL: https://www.R-project.org/>.
[5] H. Wickham and J. Bryan. usethis: Automate Package and Project Setup. R package version 2.0.1. 2021. <URL: https://CRAN.R-project.org/package=usethis>.
[6] H. Wickham, R. François, L. Henry, et al. dplyr: A Grammar of Data Manipulation. R package version 1.0.7. 2021. <URL: https://CRAN.R-project.org/package=dplyr>.
[7] H. Wickham, J. Hester, and W. Chang. devtools: Tools to Make Developing R Packages Easier. R package version 2.4.2. 2021. <URL: https://CRAN.R-project.org/package=devtools>.
[8] Y. Xie. bookdown: Authoring Books and Technical Documents with R Markdown. ISBN 978-1138700109. Boca Raton, Florida: Chapman and Hall/CRC, 2016. <URL: https://github.com/rstudio/bookdown>.
[9] Y. Xie. bookdown: Authoring Books and Technical Documents with R Markdown. R package version 0.21. 2020. <URL: https://github.com/rstudio/bookdown>.
[10] Y. Xie. Dynamic Documents with R and knitr. 2nd. ISBN 978-1498716963. Boca Raton, Florida: Chapman and Hall/CRC, 2015. <URL: https://yihui.org/knitr/>.
[11] Y. Xie. formatR: Format R Code Automatically. R package version 1.11. 2021. <URL: https://github.com/yihui/formatR>.
[12] Y. Xie. “knitr: A Comprehensive Tool for Reproducible Research in R”. In: Implementing Reproducible Computational Research. Ed. by V. Stodden, F. Leisch and R. D. Peng. ISBN 978-1466561595. Chapman and Hall/CRC, 2014. <URL: http://www.crcpress.com/product/isbn/9781466561595>.
[13] Y. Xie. knitr: A General-Purpose Package for Dynamic Report Generation in R. R package version 1.33. 2021. <URL: https://yihui.org/knitr/>.
[14] Y. Xie and J. Allaire. tufte: Tufte’s Styles for R Markdown Documents. R package version 0.9. 2020. <URL: https://github.com/rstudio/tufte>.
[15] Y. Xie, J. Allaire, and G. Grolemund. R Markdown: The Definitive Guide. ISBN 9781138359338. Boca Raton, Florida: Chapman and Hall/CRC, 2018. <URL: https://bookdown.org/yihui/rmarkdown>.
[16] Y. Xie, C. Dervieux, and E. Riederer. R Markdown Cookbook. ISBN 9780367563837. Boca Raton, Florida: Chapman and Hall/CRC, 2020. <URL: https://bookdown.org/yihui/rmarkdown-cookbook>.
[17] H. Zhu. kableExtra: Construct Complex Table with kable and Pipe Syntax. R package version 1.3.1. 2020. <URL: https://CRAN.R-project.org/package=kableExtra>.